Helicobacter pylori shows tropism to gastric differentiated pit cells dependent on urea chemotaxis - Nature.com
Abstract
The human gastric epithelium forms highly organized gland structures with different subtypes of cells. The carcinogenic bacterium Helicobacter pylori can attach to gastric cells and subsequently translocate its virulence factor CagA, but the possible host cell tropism of H. pylori is currently unknown. Here, we report that H. pylori preferentially attaches to differentiated cells in the pit region of gastric units. Single-cell RNA-seq shows that organoid-derived monolayers recapitulate the pit region, while organoids capture the gland region of the gastric units. Using these models, we show that H. pylori preferentially attaches to highly differentiated pit cells, marked by high levels of GKN1, GKN2 and PSCA. Directed differentiation of host cells enable enrichment of the target cell population and confirm H. pylori preferential attachment and CagA translocation into these cells. Attachment is independent of MUC5AC or PSCA expression, and instead relies on bacterial TlpB-dependent chemotaxis towards host cell-released urea, which scales with host cell size.
Introduction
In terms of prevalence, Helicobacter pylori (H. pylori) is a very successful human pathogen that colonizes the stomach of ~50% of the global population. Although most infected individuals remain asymptomatic, the persistence of the pathogen and the resulting inflammation are associated with an increased risk of developing gastric disease including peptic ulcer, chronic active gastritis, and gastric adenocarcinoma1,2.
H. pylori evades the luminal gastric acid by replicating in the mucus layer close to the epithelial cells3 or by colonizing the glands4. Bacteria can attach to epithelial cells by binding to Lewis antigens, frequently presented by host glycoproteins including mucin 5AC (MUC5AC)5,6, CEA Cell Adhesion Molecule 5 (CEACAMs)7,8 and β1-integrin9. Binding to integrins promotes the expression of a bacterial type 4 secretion system (T4SS), which then translocates a bacterial protein, the cytotoxicity-associated gene A (CagA), into the host cell, where it is phosphorylated by host proteins10. CagA translocation can lead to loss of cell polarity, which can allow H. pylori to obtain nutrients11,12. The bacterial T4SS and its effector CagA are thus considered the key driver of pathogenicity and a major risk factor for the development of gastric cancer13,14. Bacterial peptidoglycan and ADP-heptose induce an NF-κB inflammatory response15,16, which is thought to further drive tissue destruction and subsequent compensatory proliferation17.
The gastric epithelium is organized into flask-like invaginations, the gastric units, which are further organized into the regions pit, isthmus and gland. The pit harbours MUC5AC-expressing pit cells. The isthmus is a region of high proliferation. The gland harbours mucin 6 (MUC6)-positive neck cells, pepsinogen C (PGC)-positive chief cells, H+, K+-ATPase (ATP4A)-positive parietal cells, and chromogranin A (CHGA)-positive enteroendocrine cells. Cells of the gland can have a life span of several months18 while pit cells die within 3–5 days19,20. In vivo, it is technically not possible, to monitor adherence and CagA-translocation into the different cell types.
In vitro studies in adult stem cell-derived organoids and organoid-derived monolayers have demonstrated that gland cells mount a strong inflammatory response while the pit cells are relatively refractory to infection21,22. However, it remains unclear, whether H. pylori has a preference to attach to and translocate CagA into a particular cell type.
The recent advances in single-cell RNA sequencing (scRNA-seq) technologies allow to resolve cellular composition of tissues and organoids23. It also allows to decompose host–pathogen interaction at an unprecedented resolution, providing a powerful tool to decipher both cellular identities and their function in the complex and heterogeneous outcome of the infection24,25.
Here, we combine the power of human adult stem cell-derived gastric organoids and scRNA-seq to identify the H. pylori preferential target cell. ScRNA-seq of 3D gastric organoids and 2D monolayers derived from organoids demonstrate that the former mostly contains cells from the gland region, while the latter contains pit cells and especially a population of highly differentiated pit cells. This population is absent from 3D organoids under standard growth conditions but since adult stem cells have full differentiation capacity, 3D organoids can also be directed to generate the cell type, which is a particularly large and granular cell expressing PSCA, GKN1 and GKN2. H. pylori preferentially binds to these highly differentiated pit cells. This preference depends on the chemotaxis of the bacteria towards host urea. As a byproduct of protein metabolism, urea secretion scales with host cell size and so does the binding preference of H. pylori. Together, the data shows that H. pylori urea chemotaxis leads to preferential binding to particularly large, highly differentiated pit cells.
Results
Gastric organoids and gastric organoid-derived monolayers represent different regions of the gastric units
Organoids, as well as organoid-derived 2D monolayers, have been used as model to study H. pylori infection by us21,26 and others12,22,27. Directly comparing 3D versus 2D infection, we find different results regarding the ability of the bacteria to bind to and translocate CagA into the host cell (Supplementary Fig. 1). This prompted us to ask, whether the two systems may be more different than initially thought.
Therefore, we aimed to characterize the cell identities present in gastric organoids and 2D monolayers and compare them to freshly ex vivo isolated epithelium. To this end, we sorted freshly isolated human gastric epithelial cells based on the epithelial cell adhesion molecule (EpCAM), 3D organoids and 2D monolayers, and profiled 1241 cells from ex vivo gastric units, 713 cells from organoids and 868 cells from 2D monolayers. Of these, 1022, 602, and 696 cells, respectively, passed quality control. Unsupervised clustering using Seurat software package28 identified 7 clusters in the ex vivo gastric units (Fig. 1a, left panel) and cluster identities were assigned based on their expression levels of cell-type-specific markers (Fig. 1b; Supplementary Data 1 and 2, Supplementary Fig. 2).
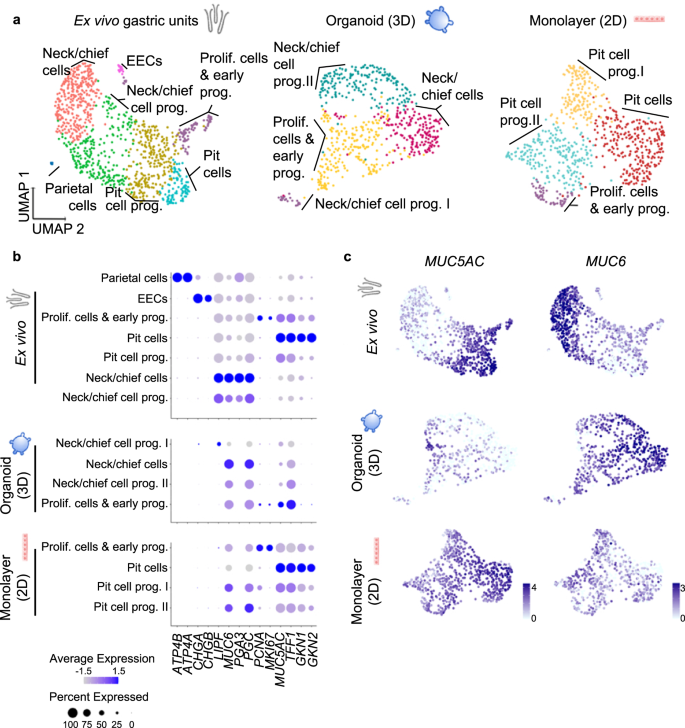
a UMAP visualisations of scRNA-seq of ex vivo isolated human gastric units (left, 1022 cells), gastric organoids (centre, 602 cells) and organoid-derived monolayers (right, 696 cells). EEC enteroendocrine cells. Cells are colour-coded according to the annotated clusters. b Dot plot showing the expression of cell type-specific marker genes within the clusters as identified in panel (a). c Expression of the pit cell marker MUC5AC and neck cell marker MUC6 colour-coded and projected on top of the UMAP visualisations as in panel a (data was normalized using 'LogNormalized' Seurat method for the expression of the gene within the dataset).
Comparing the in vitro cultured cells of organoids and 2D monolayers to the ex vivo epithelial cells, similar cell types were identified with the exception of enteroendocrine and parietal cells, which are rare or absent in organoids21 (Fig. 1a, middle and right panel). Taking a closer look, however, the two cultures were differentiated into opposing directions, which was apparent in the analysis of the main markers MUC5AC and MUC6 (Fig. 1c).
In the 3D organoids, only few cells in one cluster expressed some levels of MUC5AC, but simultaneously PGC, MUC6 and proliferation markers PCNA and MKI67, indicating that they were proliferating cells and early progenitor cells (Fig. 1a, b, Supplementary Data 1, Supplementary Fig. 2). Cells from the other three clusters expressed different levels of neck and chief cells markers such as MUC6 and PGC and were annotated as neck/chief-cells and neck/chief progenitors I and II. Gastrokine 1 (GKN1) and 2 (GKN2) were only expressed at very low levels by a few cells in the organoid sample, indicating that organoids do not contain any mature pit cells (Fig. 1b).
The analysis of 2D monolayers gave the mirror image to the organoids (Fig. 1b, c). MUC6 and PGC were expressed by one neck/chief progenitor cluster, but no mature neck/chief cells were observed in the 2D monolayers. Instead, and opposite to the organoid, the biggest cluster of cells was composed of pit cells expressing high levels of MUC5AC, TFF1, GKN1 and GKN2 comparable to the mature differentiated pit cells of the ex vivo gastric units (Fig. 1a, b, Supplementary Data 1, Supplementary Fig. 2). qRT-PCR analysis corroborated this data (Supplementary Fig. 2b).
These data suggested that gastric organoids rather resemble the gland region, while 2D monolayers resemble the pit region. We hypothesized that the initially observed different capacities of H. pylori to adhere to host cells may be rooted in the host cell identities present in the respective system and a preference of the bacteria to adhere to a specific target cell.
scRNA-seq identifies H. pylori tropism
To identify a possible target cell of H. pylori, we performed scRNA-seq of infected cells. For this, cells from human gastric organoids were seeded to form monolayers and infected with GFP-expressing H. pylori at a multiplicity of infection (MOI) of 1 or left untreated (Fig. 2a). After 6 h, cells were sorted based on GFP signal, resulting in the 3 samples "naïve" (=untreated), "infected" (GFP-positive) and "bystander" with bystander cells defined as GFP negative cells sorted from an infected sample (Fig. 2b). At this MOI, an infected sample contained usually 40% of infected and 60% of bystander cells. In total, we profiled 2170 cells (868 naïve, 870 bystander and 432 infected). Of these, 1727 cells (713 naïve, 742 bystanders and 272 infected) passed the strict quality-control thresholds (see the "Methods" section for details). The generated data from the three samples was combined and analysed using the Rare Cell Type Identification (RaceID) algorithm29. T-distributed stochastic neighbour embedding (t-SNE) projection showed that H. pylori-infected cells clustered together, while bystander cells were interspersed between the naïve cells, indicating that many of the infected cells, but not the bystander cells, differed from the naïve cells based on their transcriptome (Fig. 2c). A total of four clusters were identified based on their transcriptomic profile and annotated based on cell type-specific marker expression (Fig. 2d, e, Supplementary Fig. 3, Supplementary Data 3).
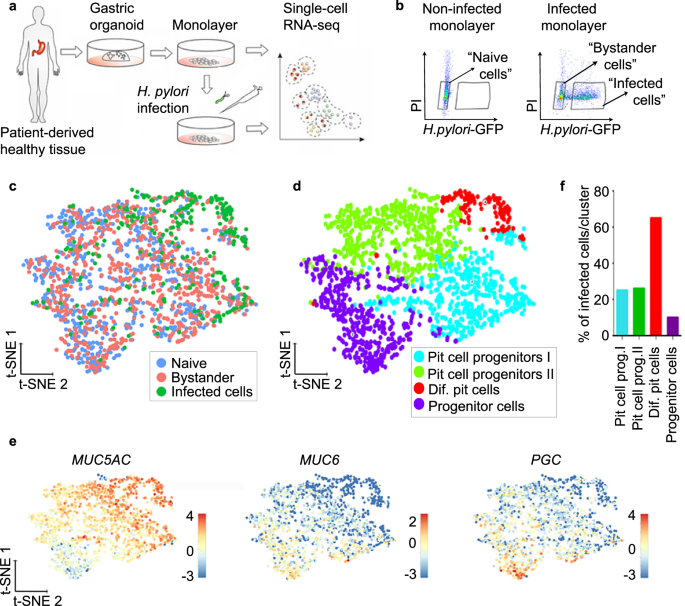
a Scheme of the experimental setup. Gastric organoids were generated from gastric healthy tissue and used to obtain 2D monolayers. Monolayers were infected with GFP-expressing H. pylori (MOI 1) for 6 h and subjected to scRNA-seq. b Representative flow cytometry dot plots of naïve and infected monolayers. Cells were gated as indicated based on GFP signal (H. pylori) and propidium iodide (PI) and subjected to scRNA-seq. c Single-cell transcriptomes from the different gates (panel b) were integrated and projected using a t-distributed stochastic neighbour embedding (t-SNE) 2D projection. The sample origin is colour-coded. d Using known gastric marker genes for the specific gastric cell types, cell identities were assigned. e Expression of known markers specific for pit cells (MUC5AC), neck cells (MUC6) and chief cells (PGC) colour-coded and projected on top of the t-SNE displayed in panels (c) and (d). f Percentage of H. pylori-infected cells per cluster as identified in panel (d).
Two clusters were composed of pit cell progenitors (clusters 1 and 2), and 1 cluster was composed of progenitor cells (cluster 3). While the smallest cluster, cluster 4, containing 8% of the total number of cells in the sample, represented the highest differentiated pit cells (Fig. 2d, e, Supplementary Fig. 3, Supplementary Data 4), it also contained the highest percentage of infected cells (65%). In contrast, in clusters 1 and 2, both representing pit cell progenitors, 25% and 26% of cells were infected, respectively. In cluster 3, representing the least differentiated cells, only 10% of cells were infected (Fig. 2f, Supplementary Data 4). Together, the data indicate that H. pylori is not restricted to, but preferentially binds to a specific subpopulation of pit cells.
The H. pylori preferential target cell is the most differentiated pit cell expressing high levels of GKN1, GKN2, CEACAM5, PHGR1 and PSCA
To further characterize this target cell population of H. pylori, we focused on the specific transcriptional profile of these cells.
Comparison of the preferential target cell (cluster 4) with the less differentiated cells (clusters 1–3) identified 19 differentially expressed genes (DEG), 5 genes with higher and 14 genes with lower expression in the differentiated pit cells (log2 ≥ 1.5, p-value ≤ 0.05) (Fig. 3a, Supplementary Data 5).
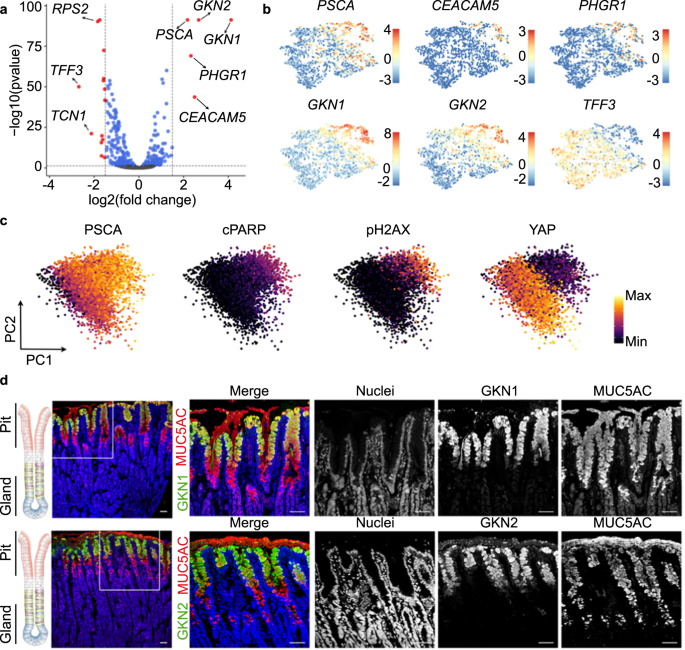
a Volcano plot showing log-fold change and −log (p-value) for the comparison between cluster 4 (H. pylori-infected enriched cluster) and the rest of the clusters (clusters 1, 2 and 3). Red dots denote DEG with p-value ≤ 0.05 and ≥1.5 log2 fold change. Blue dots denote DEG with p-value ≤ 0.05 and ≤1.5 log2 fold change. b Expression of several DEG shown in panel (a). Expression is colour-coded and projected on the t-SNE as in Fig. 2c. c Protein levels of PSCA, cleaved PARP, phosphorylated H2AX and YAP in 2D naïve monolayers measured by mass spectrometry (CyTOF). Every dot represents an individual cell and colours denote the intensity of the signal. d Confocal microscopic image of human stomach mucosa co-stained for MUC5AC (red), GKN1 (green, upper panel) and GKN2 (green, lower panel) using immunofluorescence. Nuclei were counterstained using Hoechst 33342 (blue). Scale bar: 50 µm. Data in panel c is representative results of organoid lines derived from 3 donors and 1 experiment. Data in panel d is representative of results from 2 donors and 1 experiment.
We focused on 5 highly expressed genes: GKN1, GKN2, CEACAM5, prostate stem cell antigen (PSCA) and proline, histidine and glycine-rich 1 (PHGR1; Fig. 3a). Similar to MUC5AC (Fig. 2f), the expression of these genes followed a gradient with the highest expression level in cluster 4 (Fig. 3b). Mass cytometry indicated that cells with the highest expression for PSCA were also positive for apoptosis markers (phosphorylated H2A histone family member X, pH2AX; cleaved Poly ADP-ribose polymerase, cPARP) and negative for YAP (Fig. 3c, Supplementary Fig. 4). Both suggested that the gradient indicates cellular differentiation and that the cluster 4 may constitute a highly differentiated pit cell population. To verify this hypothesis, we analysed the protein expression in histology. In agreement with previous studies30,31, GKN1 and GKN2 marked a fraction of MUC5AC-positive pit cells localized at the opening of the pit (Fig. 3d). Classical studies using autoradiography have identified these cells as the most matured pit cells32. Data from the Human Protein Atlas33 showed a similar localization for PHGR1, PSCA, and CEACAM5 (Supplementary Fig. 5). qRT-PCR analysis of these markers after infection demonstrated that their presence in infected cells was not due to the upregulation of the gene transcripts upon infection (Supplementary Fig. 6a). No bacterial binding preference was observed when using higher MOIs for infection. At an MOI of 100, H. pylori attached to all cells independent of the marker gene expression (Supplementary Fig. 6b, c).
These results indicate that H. pylori preferentially binds to a subpopulation of highly differentiated pit cells, characterized by expression of GKN1, GKN2, PHGR1, PSCA and CEACAM5, located in the upper pit region.
Directed differentiation of 2D monolayers
To verify the preferential binding for H. pylori, we aimed to control the abundance of the differentiated pit cells in the 2D monolayers, using directed differentiation as published before21. For this, standard growth conditions (EGF, Noggin, R-Spondin, Wnt, FGF-10, Gastrin and TGFβ inhibitor or in short ENRWFGTi) are modified to direct the differentiation of the cells (Fig. 4a)21. Wnt withdrawal led to differentiation towards the H. pylori target cell, as indicated by high expression of MUC5AC, GKN1, GKN2 and PSCA on RNA and protein levels (Fig. 4b, Supplementary Fig. 7a), while the addition of nicotinamide led to differentiation towards the gland cell identities as indicated by increased expression of MUC6 and PGC (Fig. 4b, Supplementary Fig. 7a). Flow cytometric analysis demonstrated that Wnt withdrawal induced an increase in size (median FSC 6.24 × 106 in ENR_FGTi_ compared to 5.10 × 106 in ENRWFGTiNi; Supplementary Fig. 7b) and granularity (median SSC 1.10 × 106 in ENR_FGTi_ compared to 7.68 × 105 in ENRWFGTiNi; Supplementary Fig. 7c), giving the impression of differentiation towards a large and granular cell, matching the expectation for highly differentiated pit cells. Analysis of PSCA staining in these conditions showed the differentiation to a PSCA+SSChigh population (37.2 ± 9.1 % of gated cells in ENR_FGTi_ compared to 10.4 ± 4.8% in ENRWFGTiNi; Supplementary Fig. 7d, e). Moreover, during the differentiation towards the pit cell lineage, higher production of mucus was observed in the culture medium compared to the control (ENRWFGTi_, Supplementary Movie 1). These results show that three types of 2D monolayers can be generated that differ in their content of pit- or gland-type cells by modifying the composition of the culture media.
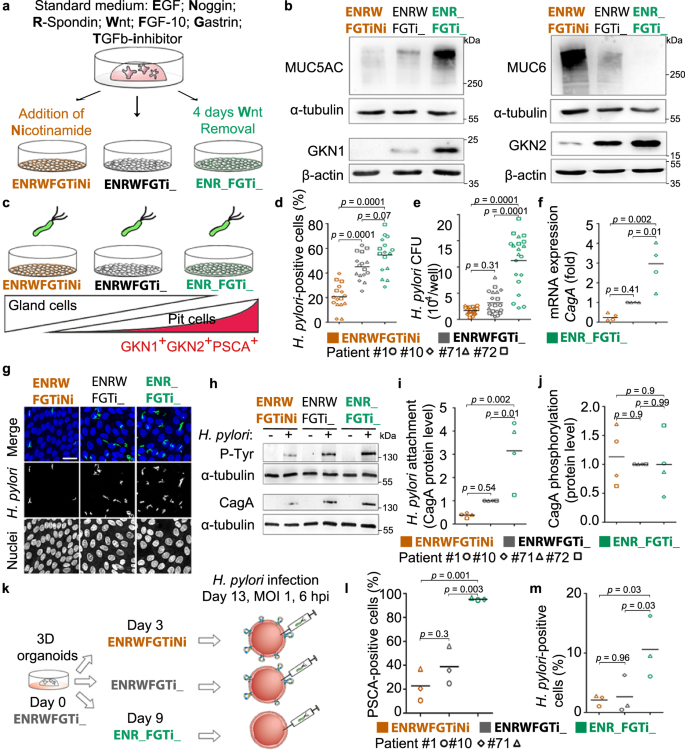
a Scheme of the directed differentiation setup. b Western blot analysis of pit markers (MUC5AC, GKN1, and GKN2) and neck markers (MUC6) in 2D monolayers upon differentiation according to the scheme in (a). c Scheme of the experimental setup. Differentiated monolayers were infected with a H. pylori strain expressing GFP at a MOI of 1 for 6 h. d–f Quantification of adhered bacteria to 2D monolayer cells by (d) flow cytometry, (e) CFU assay and (f) qRT-PCR of CagA expression (fold over host GAPDH; f). Results are normalized against standard media (ENRWFGTi_, grey symbols). g Confocal images of differentiated gastric 2D monolayers infected with GFP-expressing H. pylori. Nuclei were counterstained with Hoechst 33342. Scale bar: 25 µm. h Western blot analysis of phosphorylated CagA (P-Tyr) in H. pylori-infected 2D monolayers. i and j Quantification of bacterial attachment (CagA total protein over host alpha-tubulin; i and CagA translocation (P-Tyr over CagA; j) by western blot. k Scheme of the experimental setup. Differentiated 3D organoids were microinjected with H. pylori (MOI 1 for 6 h). l Quantification of PSCA-positive cells in 3D organoids measured by flow cytometry. m Quantification of adhered bacteria to 3D organoid cells by flow cytometry. Data from organoid lines derived from individual donors are shown as symbols (patient #1-circle, #10-diamond, #71-triangle, and #72-square), with mean values with horizontal lines. Data in panel b, d–j are representative of or show data of organoid lines from 4 donors, l and m of 3 donors, and 1 (b, f, i and j), 3 (g, h, l, m), 4 (d) or 5 (e) independent experiments. Statistical analysis of data in panels d–f, i, j and l, m was performed using one-way ANOVA with Tukey's multiple comparisons post-hoc test. Source data are provided as a Source Data file.
H. pylori adherence and CagA translocation match the presence of differentiated pit cells in differentiated monolayers
Using the three different growth conditions, we then analysed, whether the absence or presence of the target cell would affect bacterial binding and CagA translocation. For this, 2D monolayers were infected with H. pylori MOI 1 (Fig. 4c). Flow cytometric analysis showed 20.7%, 45% or 54.5% of H. pylori-positive cells in the gland cell differentiation, standard, or pit cell differentiation condition, respectively (Fig. 4d), indicating that bacterial binding increases with the presence of mature differentiated pit cells. This was matched by 1.7, 3.2 and 11.2 × 104 colony forming units (CFU), respectively, as measured by classical attachment assay (Fig. 4e). Also, expression levels of CagA indicated the similar distribution of bacterial RNA in the infected samples (Fig. 4f) and confocal imaging of the infection confirmed the higher bacterial adherence to the pit cell differentiation condition (Fig. 4g). Western blot also showed about 3-fold higher level of CagA protein in the pit cell differentiation condition. Western blot for phosphotyrosine showed that the bound bacteria also actively injected CagA into the host cells (Fig. 4h–j). To test whether other factors specific for 2D or 3D would influence binding, we used directed differentiation in 3D, which also yielded the differentiated pit cells. Flow cytometry analysis of organoids microinjected with bacteria showed that H. pylori attachment is similarly correlated to PSCA expression in 3D as it is in 2D (Fig. 4k–m).
Together, these results indicate that H. pylori preferentially binds to differentiated pit cells and that this differentiation can be directed to increase bacterial adhesion both in 2D and 3D.
Binding of H. pylori to differentiated pit cells is independent of MUC5AC and PSCA
We hypothesized that specific proteins might mediate the preference to the target cell. Candidate proteins were MUC5AC because previous studies have demonstrated the binding of H. pylori to Lewis B antigens of MUC5AC6,34 and PSCA because it is the highest expressed surface antigen-specific for this cell type in our analysis. To analyse the importance of MUC5AC and PSCA, we generated CRISPR/Cas9-mediated knockouts in human gastric organoids. Knockout was confirmed in DNA and protein analysis (Supplementary Fig. 8), but bacterial binding indicated that neither MUC5AC nor PSCA are required for H. pylori adhesion to the 2D monolayers (Supplementary Fig. 8).
Preference for pit cell depends on urea chemotaxis and cell size
We then analysed the possible role of secreted factors. To test whether gland cells would secrete bactericidal proteins to repel or kill bacteria, we incubated bacteria with supernatants from the differentiated cells. The supernatants did not have an effect, although infection did lead to the upregulation of some known antimicrobials (Supplementary Fig. 9).
To test whether pit cells would attract bacteria, we employed a classical chemotaxis assay35. All cell-conditioned media attracted bacteria, but conditioned medium from pit cells attracted about twice as many bacteria as conditioned medium from gland cells (Fig. 5a). Urea is a major chemoattractant for H. pylori36. To test whether the here observed chemotaxis was due to urea, we added 500 mM of urea, which has been shown to abrogate H. pylori chemotactic response to urea36. Indeed, the high urea abrogated the response to the pit cell-conditioned media (Fig. 5b). Deletion of TlpB, the H. pylori gene for the chemoreceptor that recognizes urea36 abrogates the preference for the pit cells (Fig. 5c). TlpB mutant bacteria also did not prefer pit cells in infections (Fig. 5d).
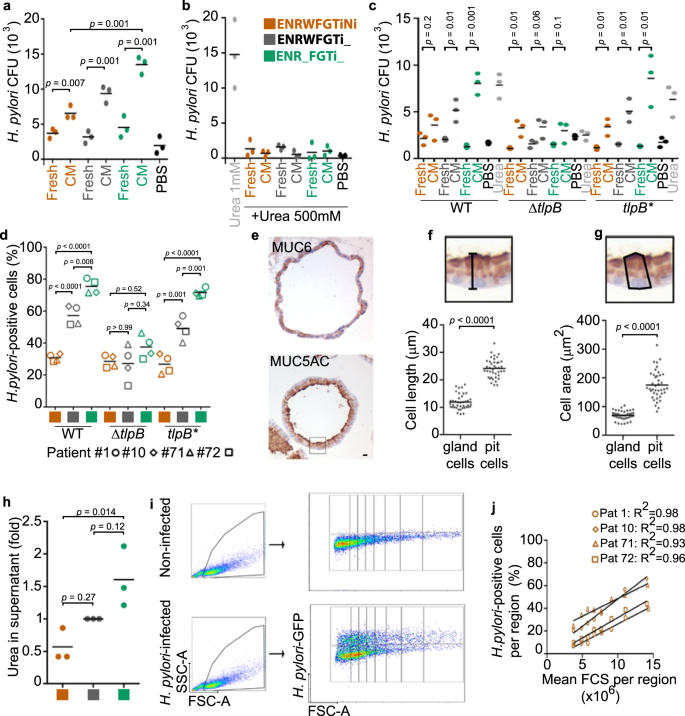
a–c Quantification of wild-type (wt, strain P12) bacterial chemotaxis towards conditioned media (a) or conditioned media plus 500 mM urea (b). Chemotaxis by H. pylori strain G27 WT, ΔtlpB and tlpB* (c). d Quantification of adhered H. pylori strains G27 WT, ΔtlpB and tlpB* to organoid-derived monolayers by flow cytometry. e Representative immunohistochemistry images of a 3D organoid stained for MUC5AC and MUC6. Scale bar: 10 µm. f and g Quantification of cell length (f) and cell area (g) of MUC5AC-(pit cells) and MUC6-positive cells (gland cells) in 3D organoids. h Quantification of urea concentration in organoid-derived monolayer conditioned media. Fold over control differentiation media (grey, ENRWFGTi_). i Representative flow cytometry analysis. j Quantification of H. pylori-positive cells in the FCS regions shown in (i). Data are shown as individual dots (panels a-c, and h, filled dot, pool of 4 organoid lines, 3 independent experiments; in panels d and j, symbols represent organoid lines derived from patient #1-circle, #10-diamond, #71-triangle, and #72-square; 3 independent experiments), mean values with horizontal lines (panel d). Data in panel e is representative of 2 organoid lines and 1 experiment. Statistical analysis of data in panels a–c was performed using multiple t-test corrected for multiple comparisons with the Holm–Sidak method; in panels d and h, one-way ANOVA with Tukey's multiple comparisons post-hoc test.; and in panels f, g, two-tailed Student's t-test.
Urea is a byproduct of amino acid catabolism. Because larger cells produce more protein37, it is likely that large cells may secrete more urea than small cells. As mentioned above, already flow cytometry analysis had shown that cells in the ENR_FRTi_ condition (pit cells) are larger than cells in ENRWFGTiNi condition (gland cells) (Supplementary Fig. 7b–e). This is also visible in the histological staining of organoids (Fig. 5e–g). Matching cells in the ENR_FRTi_ condition (pit cells) produce about 3 times higher concentrations of urea than cells in the ENRWFGTiNi condition (gland cells) (Fig. 5h). If urea secretion scaling with size would be a major determinant of H. pylori preference, this would also be visible in all differentiated cells (irrespective whether these are pit or gland cells). Indeed, flow cytometry analysis showed that percentages of H. pylori-infected cells correlate to the cell size also in the absence of pit cells (ENRWFGNiTi) (Fig. 5i, j).
We conclude that urea chemotaxis leads to H. pylori preferential binding to large cells and that the binding preference to differentiated pit cells is rooted in the cell size.
H. pylori localises in close proximity to highly differentiated pit cells in vivo
Having observed preferential binding of H. pylori to a subpopulation of highly differentiated pit cells in vitro, we examined whether this preference could be observed in vivo. For this, we used H. pylori-positive gastric biopsies from donors that underwent endoscopy diagnosis. Immunofluorescence microscopy showed that H. pylori was predominantly found next to cells expressing GKN1 at the opening of the gastric pits (Fig. 6a).
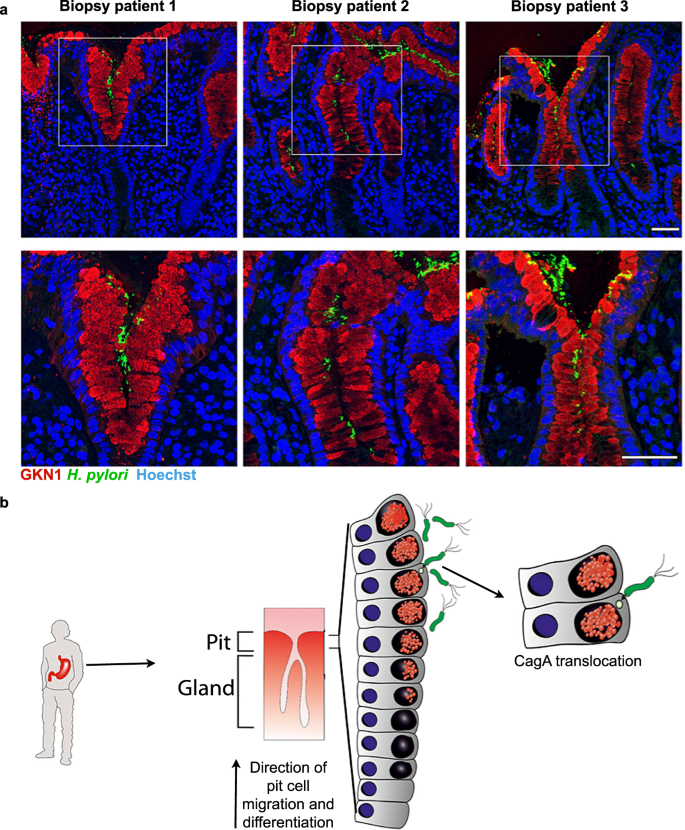
a Representative confocal microscopy images of H. pylori-positive tissue sections co-stained for GKN1 (red) and H. pylori (green). Nuclei were counterstained using Hoechst 33342 (blue). Scale bar: 50 µm. Data shown are representative images of three patients and one experiment each. b Model: H. pylori preferentially binds to and translocates CagA into highly differentiated pit cells located at the opening of the gastric units.
Overall, these observations demonstrate that H. pylori preferentially binds to highly differentiated pit cells, located in the upper area of the gastric pits, where it can translocate its CagA virulence factor (Fig. 6b).
Discussion
For H. pylori, bacterial adhesion is not only an important step for colonization in the human stomach10, but also a prerequisite for CagA translocation7,8,10. In this study, we demonstrate that H. pylori preferentially binds to a highly differentiated subpopulation of gastric pit cells. This is mirrored by high bacterial density in the pit region in patient tissue.
Already classical studies by Warren and Marshall reported that in human biopsy material, the majority of H. pylori is found close to the pit region38. This preferential location was underlined by quantification: In human biopsy material, less than 1% of bacteria are found associated with MUC6-producing gland cells39. Studies in mouse sections find more frequent colonization of the deep glands 4,40. A direct quantitative comparison between bacterial location in mouse and human infection is lacking, but the current data suggests possible differences between hosts.
Human organoids and their derived monolayers are emerging additions to the spectrum of model systems in infection biology and allow the study of human-specific pathogens with primary human epithelium. Here, a comparison of ex vivo sorted gastric epithelial cells with cells from 3D organoids and organoid-derived 2D monolayers using single-cell sequencing demonstrated that the two models represent different parts of the gastric unit: Under standard growth conditions, 2D monolayers harbour mostly cells from the pit region and their progenitors, while 3D organoids rather harbour the glandular progenitors. Both models are important for the understanding of H. pylori infection, and both can be directed towards different cell fates. Because organoids and their derived monolayers are now becoming a key model in a wide range of bacterial and viral infections12,21,22,41,42,43,44,45,46,47,48, the cell identities present should be taken into consideration when choosing the experimental system and interpreting results.
In this study, we have characterized the highly differentiated pit cell as a preferential target cell for H. pylori binding. This subset of pit cells is characterized by expression of high levels of MUC5AC, GKN1, GKN2, CEACAM5, PSCA and PHGR1. MUC5AC contains Lewis B antigens, which are known to be bound by H. pylori adhesins. PSCA is a membrane-anchored glycoprotein expressed in several organs including the stomach. Its function has not been elucidated yet, but genetic variants of this gene have been associated with gastric cancer49. However, in our experiments, MUC5AC or PSCA expression was dispensable for the binding. CEACAM5 is a receptor for HopQ, an H. pylori adhesin7,8. In our sample, CEACAM5 transcript was only detected in a fraction of the target cell population and thus it is likely that this is not the only factor important for bacterial binding in our setting. Instead, we found that chemoattraction to the host cell metabolite urea underlies the preferential binding to differentiated pit cells. Urea attraction has been described early using capillary tube assays50,51. Recent work has identified that H. pylori directly senses urea via the chemoreceptor TlpB36. TlpB has an unusually high affinity for urea and H. pylori can detect concentrations of as low as 50 nM. Simultaneous degradation of urea by H. pylori-secreted urease prevents the receptor from being saturated. This highly sensitive urea-sensing by TlpB is important for the long-term colonization of the stomach36. Interestingly, TlpB gene expression is regulated via targeting of a variable G-repeat in its sequence by the small regulatory RNA RepG52, which was also recently shown to regulate HP0102, a gene involved in LPS biosynthesis53.
The importance of cell size, for example in aging and stem cell capacity, is only beginning to be discussed54,55. Here we show that the urea sensing of H. pylori leads to a bacterial preference for larger cells. Notably, large size characterizes senescent cells56,57. Our size measurements in organoids correspond well to measurements in human organoid-derived monolayers22 and mouse tissue, with heights for gland cells from 13 µm to 19 µm, and heights for pit cells of 20–45 µm, with the length of the pit cells increasing with differentiation19,58. Notably, pit cells are the cells with the highest turnover and are being replaced about every 3 days59. In addition, contrary to gland cells, pit cells are refractory to mounting an inflammatory response to infection with H. pylori21. Thus, with this very sensitive chemotaxis towards urea, evolution has shaped a fine-tuned system, in which bacteria target those cells that are not only mounting the lowest immune response but, are also replaced most frequently in the epithelium and thus most dispensable for the tissue.
Methods
Gastric organoid generation and organoid-derived monolayer seeding
Human gastric tissue was obtained from four donors (age range 32–82 years) that underwent partial gastrectomy at the University Hospital of Würzburg. This study was approved by the ethical committee of the University of Würzburg (Approval 37/16) and informed consent was obtained from all the donors.
Generation and culture of gastric organoids were performed as described before21,60. For propagation, organoids were split every 12 days at a ratio of 1:10 using mechanical splitting. Culture medium was replaced every 2–3 days. Propagation culture medium contained: Advance Dulbecco's modified Eagle medium/F12 (12634028, Thermo Fischer Scientific) supplemented with 10 mmol l−1 HEPES (15630056, Thermo Fischer Scientific), 1x GlutaMAX (35050-038, Thermo Fischer Scientific), 1xB27 (12587010, Thermo Fischer Scientific), N-Acetylcysteine 1 mM (A9165-25G, Sigma-Aldrich), Noggin conditioned medium 10%, R-spondin1 conditioned medium 10%, Wnt conditioned medium 50%, EGF 50 ng ml−1 (AF-100-15, Peprotech), FGF10 200 ng ml−1 (100-26, Peprotech), Gastrin 1 nM (3006, Tocris), TGFβi 2 µM (A-83-01, Tocris), Primocin 100 ng ml−1 (Ant-pm-1, Invivogen). The propagation culture medium was named ENRWFGTi_. After seeding RHOKi 10 µM (Y-27632, Sigma-Aldrich) was added.
Gastric organoid-derived monolayers were generated from 8-day-old organoids. Organoids were collected and mechanically disrupted. Organoid fragments were washed with Advanced DMEM/F12 containing GlutaMAX 1x (35050-038, Thermo Fischer Scientific), 10 mM HEPES (15630056, Thermo Fischer Scientific) and centrifuged at 400×g for 5 min. Organoid fragments were resuspended in TrypLE Express (12605028, Gibco) and incubated at 37 °C for 10 min. Single-cell suspension was washed and centrifuged at 400 × g for 5 min. Single cells were counted and seeded at 15,000 cells/well in 48-well plate (833923, Sarstedt) using the propagation culture media (ENRWFGTi_). After seeding RHOKi 10 µM (Y-27632, Sigma-Aldrich) was added. Monolayers were allowed to grow and differentiate for 14 days to reach 90–100% confluency. Six days prior to infection, Primocin was exchanged for H. pylori compatible antibiotics (vancomycin, nystatin and trimethoprim, concentration as described for the bacterial cultures).
To direct the culture toward the gland cell differentiation, nicotinamide 10 nM (N0636, Sigma-Aldrich) was added to the culture media on day 4 (for organoid-derived monolayers) or day 3 (for 3D organoids; ENRWFGTiNi). To induce differentiation into pit cells, Wnt was removed from the culture medium on day 10 (for organoid-derived monolayers) or day 9 (for 3D organoids; ENR_FGTi_).
Bacterial culture and infection
The clinical isolates H. pylori strain P12 and its derivate expressing GFP constitutively from a chromosomal locus were previously described61 and it was a kind gift of Thomas F. Meyer. The H. pylori isolate strain G27, ΔtlpB and tlpB complemented strain (tlpB*) were previously described36 and a kind gift from Manuel Amieva. We also received G27 and tlpB mutant from Cynthia Sharma52 and this showed the same results. H. pylori was grown on GC agar plates (CM0367B, Oxoid) containing 10% of heat-inactivated horse serum (S0900-500, Biowest) and supplemented with nystatin 0.2 µg ml−1 (N3503, Sigma-Aldrich), trimethoprim 0,25 µg ml−1 (T7883, Sigma-Aldrich), vancomycin 1 µg ml−1 (0242.3, Carl Roth) and kanamycin 8 μg ml−1 (C0378, Sigma-Aldrich). All bacterial cultures were incubated under microaerobic conditions (85% N2, 10% CO2, 5% O2) at 37 °C and in the case of liquid culture with orbital shaking (140 rpm).
For infection, bacteria were grown on plates for 72 h, before passaging to a fresh plate and let to grow for an additional 24 h. A day before infection, scrapes from agar were used to inoculate 10 ml of brain heart infusion (BHI) broth supplemented with 10% heat-inactivated FBS (S0615/1109D, Merck Millipore) and antibiotics for 12 h. This culture was used to set up an overnight culture (14 h) for the infection. For this, the 12 h H. pylori culture was diluted to an OD600 of 0.05 in BHI broth and grown at 37 °C under microaerobic conditions (85% N2, 10% CO2, 5% O2) with shaking until OD600 0.5 (14 h). Bacteria were harvested by centrifugation and resuspended in the organoid culture medium. Bacteria were then added apically onto the organoid-derived monolayers with an MOI of 1 for 6 h unless otherwise indicated. After 6 h incubation, unbound bacteria were removed by 3 washes with PBS and cells were collected and further processed for western blot, flow cytometry, RNA isolation, microscopy, or scRNA-seq as de...

Comments
Post a Comment